Microbes are some of the most abundant organisms on this planet. They are everywhere and in everything! Microbes are found in places like deep underground mines, at the bottom of the ocean, suspended in polar ice caps, and in your digestive tract.
Microbes include bacteria, archaea, some single-celled fungi, and viruses. Fungi and bacteria are self-sufficient living things and use chemicals or sunlight for food. But viruses are different. Viruses need another living organism to use as a host for replication and survival. There are viruses that can infect any living organism, even other microbes. A group of viruses known as bacteriophages (or just phages) infect bacteria. Phages take over bacteria and rewire their molecular machinery to steal precious resources. The infected bacteria are then forced to assemble an army of phages until the cell bursts.
These bacteria-popping viruses, known as lytic phages, can do more than infect bacteria. It turns out phages can also be used to discover important past atmospheric compositions, because there are viruses hidden in the polar ice caps. These viruses and microbes are perfectly preserved in the ice, allowing scientists the chance to identify how they survived and what Earth’s atmosphere was like when they were alive.
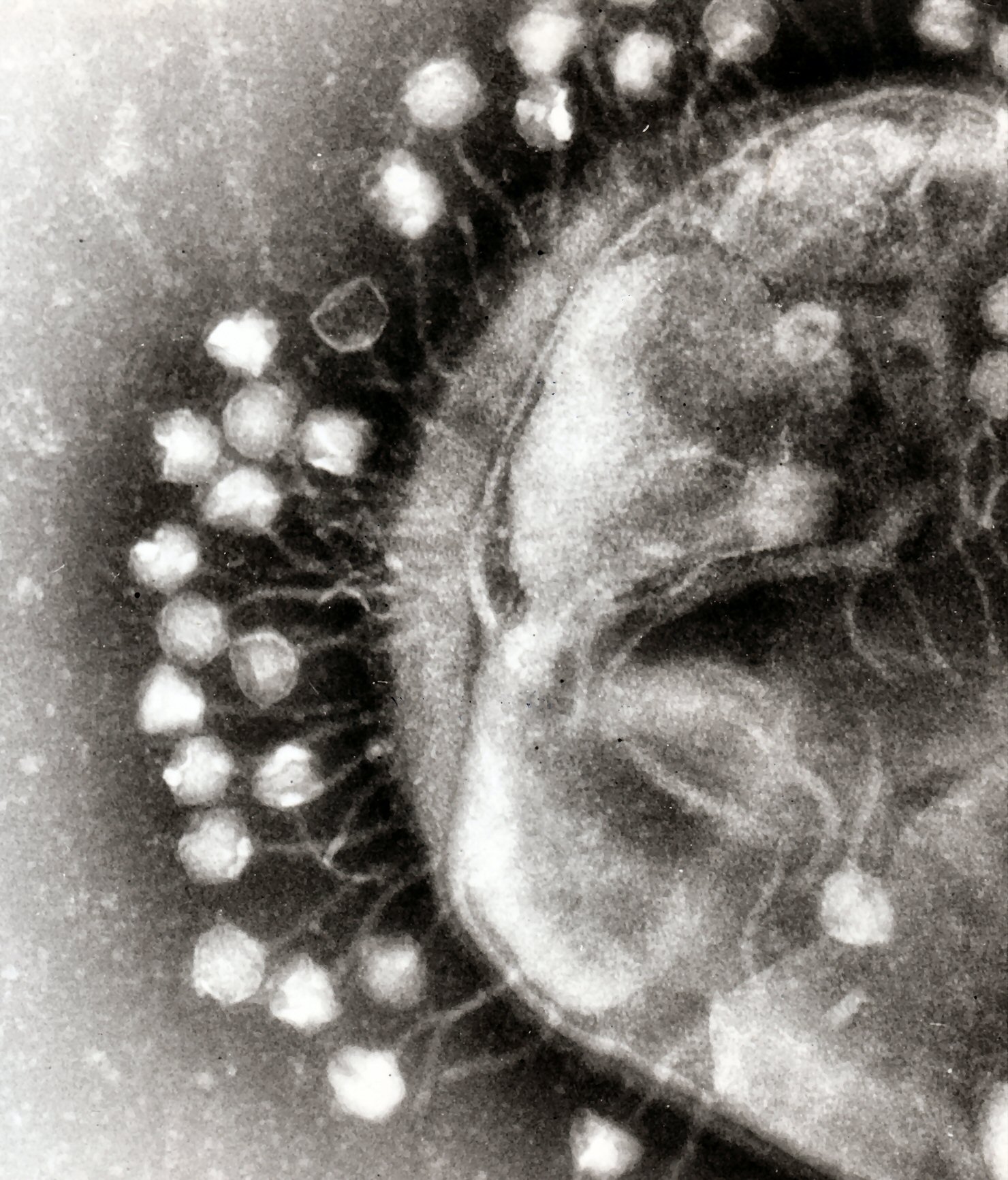
Bacteriophages on a cell membrane before infection. Dr. Graham Beards, CC BY-SA 3.0
As temperatures rise, more of the Earth’s ice supply melts and is released into the oceans. This melted ice water contains ancient microbes that could impact our current ecosystems. To learn more about what could be lurking in the ice, a group of microbiologists from Ohio State University drilled out two long cylinders of ice, called cores, from the Guliya ice cap of the Tibetan Plateau. One core was shallow, around 34.5 meters (113.18 feet) deep, and the other deeper, at 51.86 meters (170.14 feet) below the ice. These cores contained ice that ranged from 71 to 14,400 years old.
One of the toughest challenges when working with ice cores is that they are prone to contamination from other microbes not found in the core. The group attempted to solve this problem by covering artificial cores in a known mixture of bacteria, phages and free DNA. They cleaned the cores by cutting and washing them using several steps. First they cut 0.5 centimeters off with a saw, then they washed the core with ethanol and water to clean off any remaining contaminants. Finally, they tested the cleaned cores for remnants of the known mixture and found no trace of contaminants. The group states that the new cleaning method can be used to prevent any possible contamination introduced during drilling and sample handling.
Next, they sliced each core into pieces corresponding to different depths. Scientists then extracted DNA from these samples, and sequenced the DNA in order to identify what bacteria are present. Among the many species hidden in the ice cores, one called Methylobacterium was common to all of the samples. Methylobacterium uses methane for energy, and is widespread on Earth in water, air, and soil. And, along with Methylobacterium were the phages that infect it!
The presence of methylobacterium in the ice cores hint that both the bacteria and phages could be of soil, plant or air origin.

Scientist showing off an ash layer in an ice core. Source: Dargaud, CC BY-SA 4.0
Around 42.4% of the phages were identified as temperate phages. Unlike lytic phages that destroy the cell, temperate phages will infect bacteria and become one with the bacteria’s DNA.
The scientists determined that temperate phages are more desirable than lytic when phages undergo freezing conditions. Temperate phages can survive safely within the host after being frozen, and upon being thawed they can convert back to lytic to begin replication.
The microbiologists hypothesized that these temperate phages could change the shape or function of their host bacteria. They discovered four genes that the viruses could pass on to their host to change its metabolism or cause it to develop a propeller-like structure called a flagellum. Two genes were found to interfere with carbon cycling or sulfation for electron transfer. The other two could be used for flagella construction to increase host nutrient uptake.
Understanding how phages interact with their host bacteria allows scientists to observe how ancient microbes could have interacted while in communities, and predict how current microbes and phages react to extreme conditions. The biggest problem with ice core processing is contamination inhibiting microbial DNA recovery. The decontamination method the group developed will make ice core archiving easier. With this new method, the scientific community can access more ice cores for research and more viruses can be discovered.
As the ice caps melt, there is reason to suspect that ancient bacteria and viruses will be released into the oceans. Ice core archives help scientists understand what traits, beneficial or harmful, might be released into the environment as a result, and how these new traits will change the ocean ecosystem. It is important to study these microbes before they are released because they can provide an insight into the atmospheric and environmental compositions at the time they were deposited into the ice. We can use this information to predict how our current climate may be affected by these microbes.


